Antiviral Therapies: Active Nucleotid Analogs
We also selected a few modified purine analogs including modified adenosines, inosine and modified guanosines. Many of these modifications interfere with conventional Watson-Crick hydrogen bonding to the complementary bases. Furthermore, "deviant" base pairs can also contribute mutations: reverse Watson-Crick, wobble, syn-anti (purines), tautomers (enol, imino), ionized, Hoogsteen, reverse-Hoogsteen base pairs. Our nucleotide analogs are both of natural (in vivo) or synthetic (ex vivo) origin.
Active nucleotide analogs stabilizing viral quasi-species strains are almost exclusively purines, mainly modified adenosines.
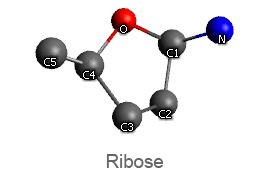
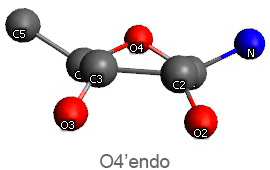
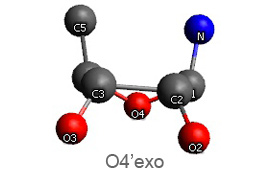
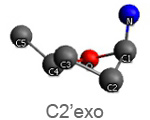
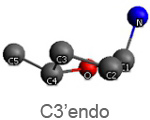
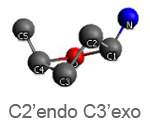
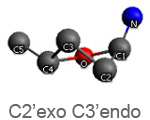
Active nucleotide analogs affecting polymerase processivity exhibit in general modifications in their ribose moiety, mainly controlled by the ribose pucker(1).
Endo and exo puckerings have dramatic consequences on the distance between N attached to C1’ and C5’ (for instance maximal for C3’endo, minimal for C3’exo, maximal for O4’endo, minimal for O4’exo (about half that of O4’endo). In addition, the puckerings stretch, or to the contrary cripple, the N-C1’-C4’-C5’ conformations, with dramatic consequences on the steric positioning of C5’ and its triphosphate of the last nucleotide attached to the nascent chain.
Since polymerases interact with the phosphate backbone and the minor groove of the RNA, their binding is largely mediated by electrostatic interactions between the RNA and the "thumb" and "palm" domains of the metaphorically hand-shaped viral polymerases. When advancing along the RNA template after adding a nucleotide, the interactions of the polymerases with the minor groove dissociate, but those with the phosphate backbone remain. The stretched puckering mentioned above may force the C5’ bound triphosphate group to bend away from the catalytic polymerase center, eliciting abortion of the elongation process.
We have many active nucleotide analogs with selected ribose puckering or puckering trends, both with, or without, modified bases. Our patented principles allow to design many more...